First single page app
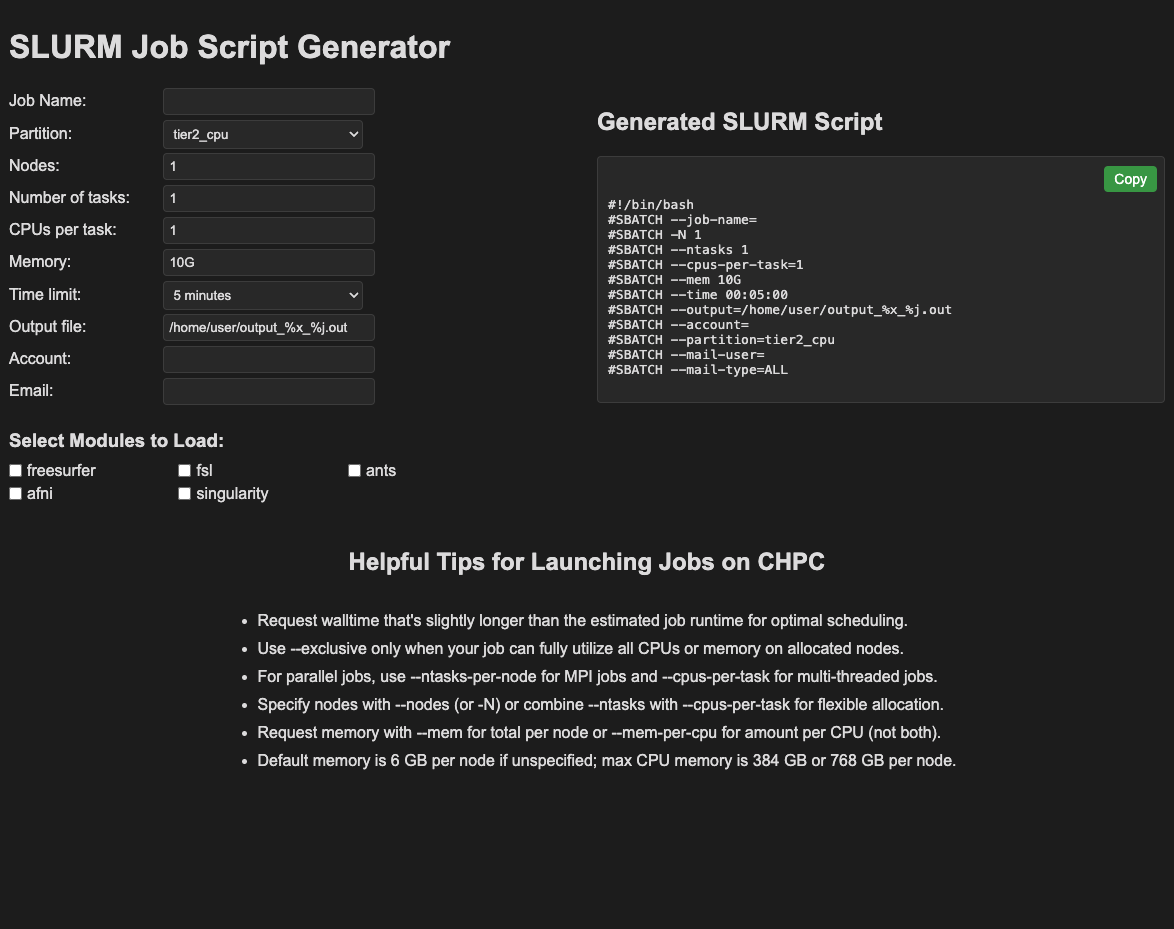
Realizing that some of my labmates were interested in using our computing cluster for their own MRI data analysis, I decided to make a single page app to help them set up the preamble for their SLURM job scripts (you can find it here). I find that people tend to have difficulty remembering what all the SBATCH options are, so I thought it would be helpful to have a simple tool that would guide them through the process. When people are learning about neuroimaging preprocessing, they often have a bunch of other technical information to keep in mind. For many people, using the command line on the cluster is sort of foreign, bash scripts are not as common as they used to be, and thinking about job scripts is yet another thing to keep track of. Right now I also hard coded in a list of software modules that are available on the cluster, but in the future I would like to make this more dynamic.
The app
The app is built with simple HTML, CSS, and JavaScript. It allows users to select their partition, specify number of nodes and memory per core as well as select a list of software modules to load (from a pre-defined list that is usually used in our lab).
Next steps
For this app, I’d like to see how much labmates actually end up using it. So far there aren’t many people in the lab who use the cluster, but demand will increase soon as more people start scanning. The other thing is that I currently hard coded in the list of software modules as well as the look up table for the time, memory, and node limits per partition. In the future I would like to make this more dynamic. Maybe by pulling from the website of the computing core at Wash U.
In addition to the app, I also learned how to deploy it on a digital ocean droplet using dokku. I may start posting some of my experiments on the droplet in the future in case people want to try them out. The eventual goal though is to host dynamic explanations on the droplet and link to them from my main site here.